Protein dynamics and disorder
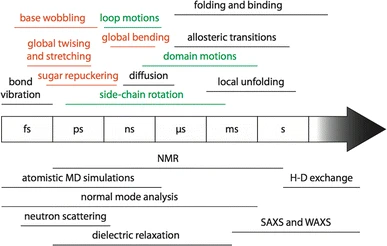
Although we often visualise proteins in terms of a static structure, all proteins undergo motions on a wide range of timescales, many of which are functionally important. NMR provides tools for measuring these dynamics in both folded and intrinsically disordered proteins. A number of applications are discussed below.
Backbone flexibility
There are several NMR methods that give information on backbone flexibility. As discussed in the pages on 1D experiments and 15N-HSQCs, you can tell if a protein is globally disorded by looking at the distribution of chemical shifts. To get more detail on specific residues, you can record variants of the 15N-HSQC experiment that record the relaxation properties of amide 15N nuclei along the backbone. These relaxation properties can be combined to estimate the timescale and amplitude of motions along the backbone, using model-free analysis and software such as Relax.
A complement/alternative to 15N relaxation studies is to predict the backbone flexibility from the carbon chemical shifts measured during the backbone assignment process. Secondary structure causes characteristic changes in the carbon chemical shifts, while flexibility tends to move them towards average "random coil" values. Programs such as TALOS-N can use these shifts to estimate the S2 order parameter, which is correlated to the amplitude of motions along the backbone.
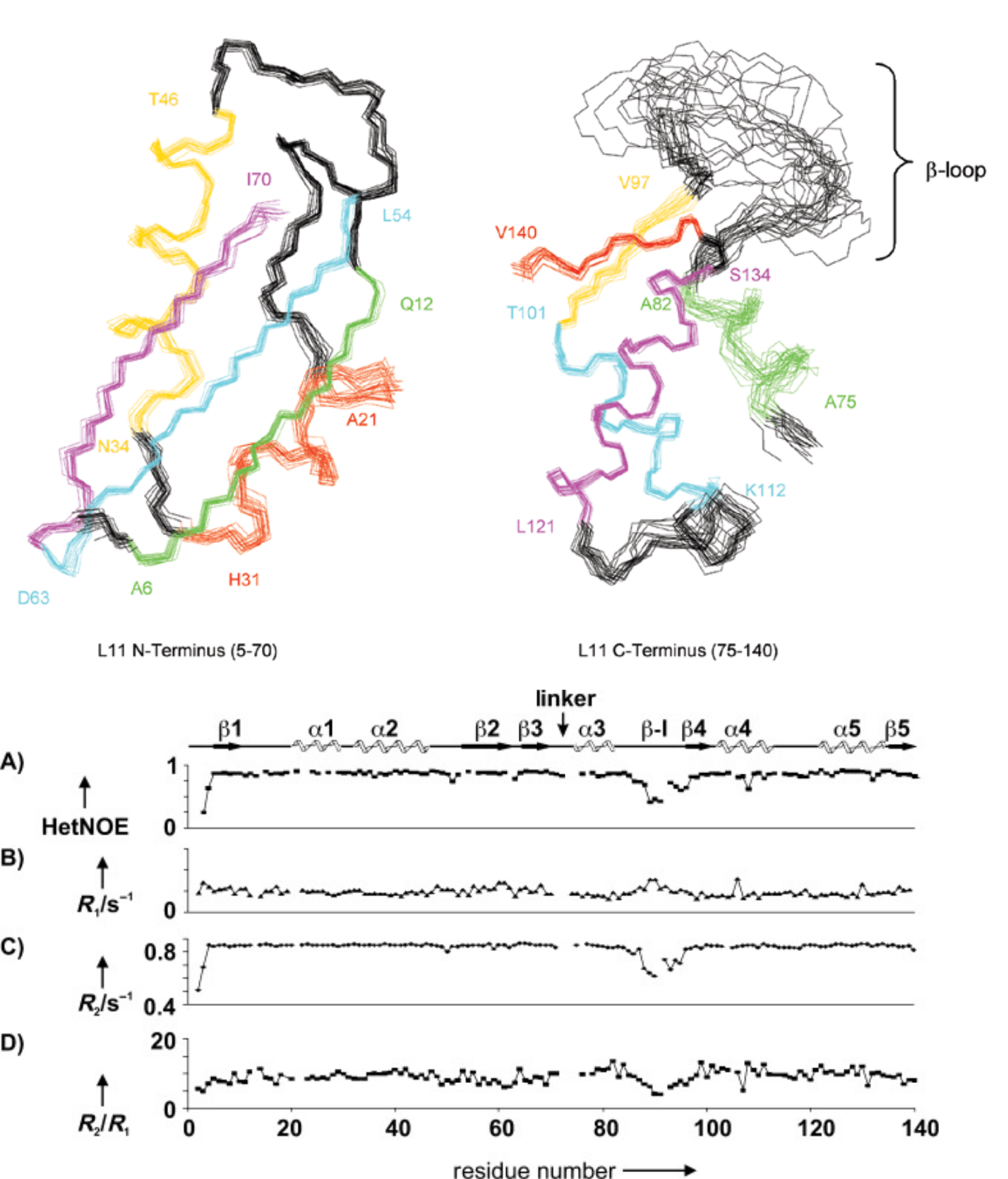
Transitions between conformations/oligomeric states
Some processes cause larger changes to the entire protein or to a subregion: these include global changes in conformation (eg transitioning between open and closed forms) or formation of oligomers or complexes. These transitions usually result in change in peak position and/or intensity in the 15N-HSQC. The exact appearance depends on the rate at which the protein transitions between the states: for processes which are fast on the NMR timescale (typically kex > 100-1000 /s), the peak will move to the weighted average of the two states; while for slow processes (kex < 10-100 /s), seperate peaks may be observed for the two states, and information on exchange rates extracted using EXSY experiments. (There is unfortunately an intermediate regime where signals tend to disappear entirely, showing that an exchange process is occurring but not revealing its details - this typically happens when kex is roughly 100-1000 /s).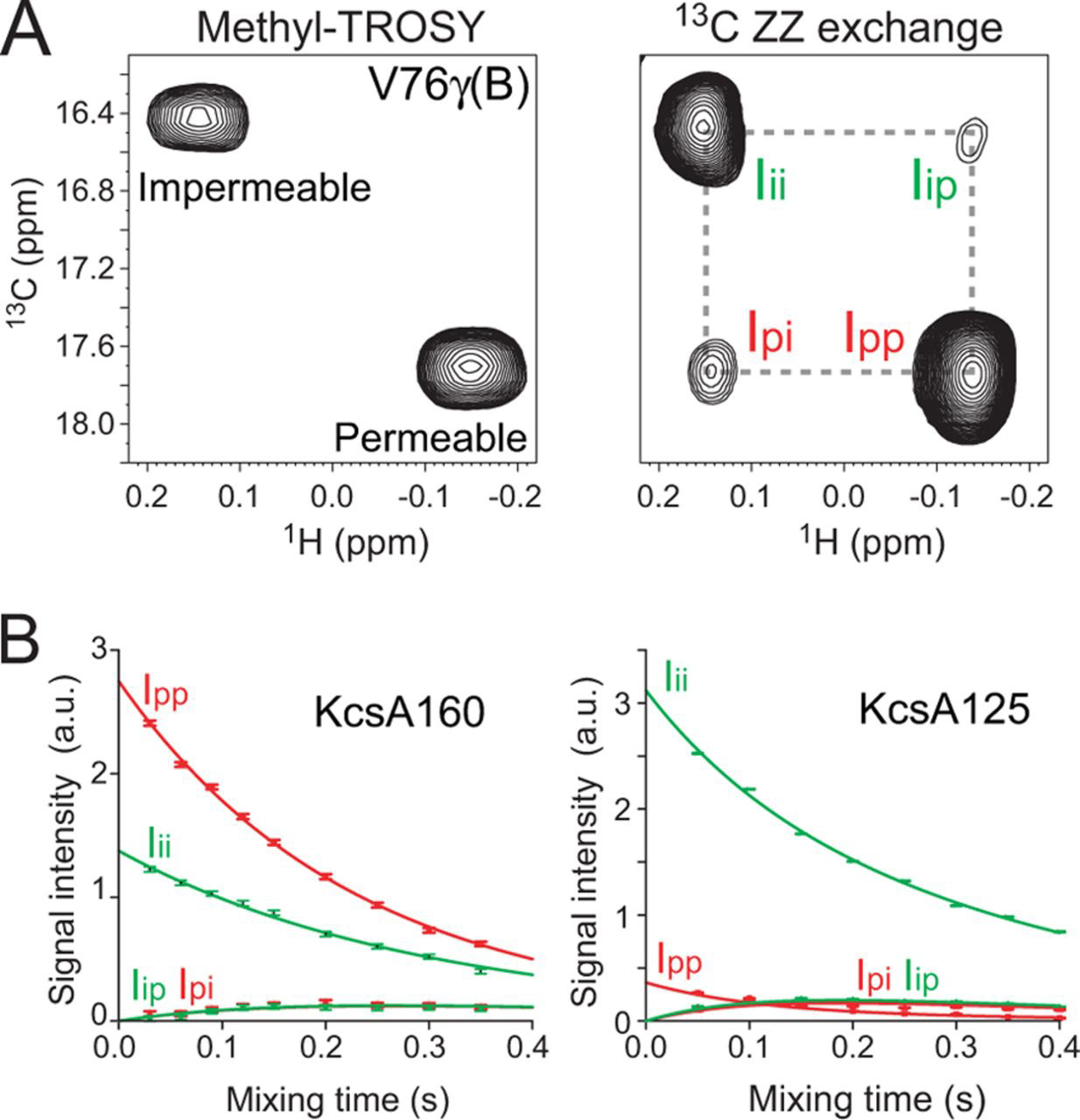
Note on timescales in NMR
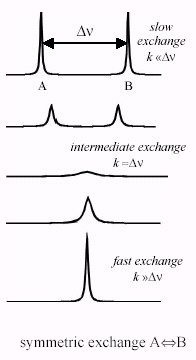
We can categorise dynamic processes as fast, intermediate or slow with respect to the NMR timescale, but what do we mean by this? As an example, consider two protein states that can exchange, and focus on one atom that is different between them. Because this atom is in a different chemical environment in the two states, it will have different chemical shifts. Although we usually measure chemical shifts in ppm compared to a reference, they really represent the precession frequency of the nuclear spin, and can be measured in Hz.
The appearance of the NMR spectra depends on how the frequency difference between the states compares to the rate of exchange between them. If the exchange rate is much slower, you observe two separate peaks in the spectrum. On the other hand, if the exchange rate is faster than the frequency difference then you observe a single sharp peak at the average frequency. When the exchange rate is similar to the frequency difference, a single broad peak is seen.
To give a concrete example, suppose we are looking at a proton which differs in chemical shift by 1 ppm between the two states: on our instrument, that corresponds to a frequency difference of 700 Hz, or a timescale or 1-2 ms. Exchange processes that are much faster than this (such as exchange between conformers in an intrinsically disordered protein) will give rise to a single peak, while exchange processes that are much slower (such as dissociation of a tightly bound ligand) will give rise to two separate peaks.
Transitions to rare/excited conformations
Sometimes the interesting state of a protein isn't the most populated state, but is instead a transient, excited state. This can be the case for multi-step enzyme reactions, or for proteins which are regulated by a switch between active and inactive states. These transient states are typically too low in concentration to be observed directly, but several NMR techniques can indirectly detect and characterise them through observation of the more populated state. These techniques include Chemical Exchange Saturation Transfer (CEST) and relaxation dispersion, and are suitable even for systems in intermediate exchange.
Side chain dynamics
13C nuclei can be used as probes of side chain dynamics in the same way that 15N nuclei are used to probe the dynamics of the backbone. Similar experiments can be used: relaxation measurements to probe fast motions, and chemical exchange and relaxation dispersion to probe slower motions. These methods are particularly applicable to methyl groups or aromatic side chains. The main caveat is that the side chains must typically be isotopically labelled only at specific sites (to avoid interference from J coupling between neighbouring carbons), which requires the use of expensive specifically-labelled amino acids in the growth medium.
Requirements
Most of the dynamics experiments above require 15N labelled protein at a reasonably high concentration (at least 200 uM, but preferably higher). 13C labelling is not necessary in most cases, although may be needed for more specialised experiments, or ones which look at side chain dynamics.